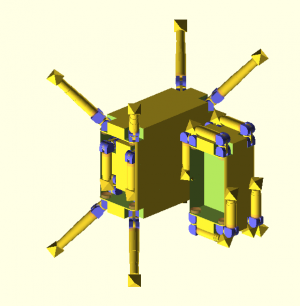
Molecular assembler (disambiguation)
Note: The concept of advanced molecular assemblers for diamondoid materials is outdated!
The current concept for advanced productive nanosystems of the "in-vacuum gem-gum technology" type are atomically precise small scale factories.
The three main problems with molecular assemblers are that they are:
- inefficient
- hard to reach
- undesirable
But molecular assemblers are:
- not fundamentally impossible
Molecular assemblers are member of the class of mobile naoscale robotic devices ("nanobots").
Molecular assemblers may emerge as products of advanced nanofactories (among other more useful "nanobots").
Contents
- 1 The idea
- 2 The ribosome and similar artificial biomimetic nano"machines"
- 3 Block placing assembler linkage
- 4 Possible exceptions
- 5 Microcomponent mainenance units ≠ Molecular assemblers
- 6 Molecular assemblers in the technical book Nanosystems
- 7 Related
- 8 External links
The idea
The idea is/was to create a machine with side-lengths of a few hundred nanometers which packages all the functionality to produce useful products and also make copies of itself (directly with diamondoid mechanosynthesis). This way you get an exponential rate of replication and can produce macroscopic goods in reasonable amounts of time.
Reasons for the three main problems
inefficiency:
It turned out that packaging all the functionality into such a small package is a rather unbalanced and inefficient approach for in-vacuum gem-gum technology. This can be seen in the nanofactory cross section image where it is visible that the bottommost assembly levels (there arranged as stacked coplanar layers) take the largest portion of the stack. In the small package of an assembler the bottommost layers would be highly underrepresented making it rather slow (and inefficient).
difficulty to reach:
See page: "Direct path".
reason for undesirability:
The grey goo horror fable toned waay down to realistic levels.
Still far down the road in the future (state 2017 .. 2022) and heavily limited by the requirements of the reproduction hexagon or replication pentagon.
Why nanofactories are more efficient than molecular assemblers
Consequences of lack of space
Molecular assemblers come with very little internal space. This has several detrimental consequences.
Unlike molecular assemblers nanofactories allow for highly efficient semi hard-coded mass production of standard parts.
Semi hard-coded nanorobotics allow for a much increased spacial reaction site density.
The total throughput results from the product of
- spacial reaction side denstity and
- temporal reaction frequency
In molecular assemblers temporal reaction frequency can't be used to compensate for lower spacial density as friction losses grows quadratically with speed and thus relatively low speeds are (~5 mm/s) are proposed. Bearing area per reaction site assumed to be the same. Furthermore bigger assembly chambers also mean lower frequencies for the same speed.
Standard part mass production may seem like a big restriction but it need not be.
- There will be many standard crystolecules of the same type needed like e.g. standard bearings
- Semi hard-coded structures can be reconfigured. It just needs disassembly and reassembly
- Software will likely be capable to auto-compile a crystolecule design into a specialized assembly-line for this part
- For very few special parts a way slower general purpose cell is still desirable (not a molecular assembler though)
Other benefits of convergent assembly (<- as listed on the page) cannot be reaped too as there's not enough space for higher assembly levels.
Consequences of maximally replicated replication capability
Every molecular assembler needs to carry around stuff for it's replication capability.
The code may be factored out in some models but there is still
a lot of redundant stuff that is just needed for self replication.
This could be called the "replication backpack overhead".
Due to the premise of molecular assemblers being very compact there is a lot
(pretty much maximal) redundancy and associated overhead.
Necessity of product production-device separation
Volumetric scaffolds of molecular assemblers makes separating
the product from the assembler crystal (or soup) way more difficult than in a nanofactory chip.
Especially when solid products are desired fractal like last remaining removal channels
suffer from extreme slowdown. (wiki-TODO: add link to existing relevant page)
Going against natural scaling laws
Due to the scaling law of higher throughput of smaller machinery
only a very small volume suffices for practical levels of throughput.
Filling a whole volume with nanomachinery would lead to uselessly high throughput.
Well, ok, molecular assemblers sabotage themselves in throughput due to
the aforementioned unnecessarily big volume-per-reaction-site.
A thin chip is the natural choice for a highly optimized (target tech) system.
It provides sufficient productive volume and also is easy to cool due to high surface to volume ratio.
Assembler hype hiding progress to nanofactoies
The assembler concept was a natural and obvious bioanalogy to introduce initially.
Continued refinement with exploratory engineering quickly led away from it though but this went almost unnoticed.
The combination of their appearance (legs or other mechanisms to move about) with their very tightly packed capability of self replication in their vacuum "belly" that seem akin to a "whomb" was one factor in leading to the situation that the public started to perceive this technology as swarms of tiny life like nano-bugs that could potentially start uncontrollable and unstoppable self replication.
Why this is a rather miss-informed perception can be read up here and here.
- Dystopian SiFi fantasy anchored the idea of assemblers in the public perception (at least in USA and UK).
- Nanofactories coming close to the newer detailed concepts remain yet to be seen in fiction.
Partial design aspects of molecular assemblers that remain applicable
Many considerations about assemblers are still relevant:
- methods for movement e.g. for the transport of microcomponents and self repair by microcomponent replacement in the higher assembly levels of nanofactories. The legged block mobility design is also known from the concept of (speculative) utility fog but has other design priorities in a manufacturing context like more rigidity and less "intelligence".
- methods for gas tight sealing and locking parts out
- and many more ...
- the design of robotic mechanosyntesis cores
Old assembler designs
Quite a bit of thought was put into the assembler model (wiki-TODO: link relevant parts in KSRM). Either they where supposed to swim about in a solution or there was some form of movement mechanism in a machine phase scaffold crystal envisioned like:
- sliding cubes (wiki-TODO: TODO add references)
- legged blocks (wiki-TODO: TODO add references)
The ribosome and similar artificial biomimetic nano"machines"
These are also often called molecular assembler although they:
- are not capable of independent self replication
- are critically dependent on brownian motion / diffusion transport
- can only assembles floppy linear chain molecules which again need brownian motion to fold into something useful
Block placing assembler linkage
Unlike "diamondoid molecular assemblers" this idea is not outdated.
Atomically precise building blocks from structural DNA nanotechnology that are pre-produced by self assembly
could be assembled to passive block manipulator linkages by those same passive block manipulator linkages after a first one was put together manually.
Actuation could be from a chips surface (see technology level I) and self replication could work in the form of exponential assembly.
- Crystolecule assembly robotics might become capable of more or less compact self replication with predelivered "vitamin" pasts from the mechanosynthesis cores.
- Wikipedia: Clanking replicator. A term to distinguish macroscale selfreplication from nanoscale selfreplication. But crystolecule level self replication is very similar to macroscale self replication. So the meaning can be dragged back. A clanking nano replicator so to say.
(Sidenote: Actual clanking "sounds" should be avoided. Sound emission = loss of energy = inefficient operation = need for waste heat removal)
Possible exceptions
Occasions where somewhat molecular assembler like designs may not yet be completely outdated.
The only place where the slow and inefficient molecular assembler concept may be practically usable is for
self repair situations where the demand on product throughput rate is exceptionally low.
Like fixing low rates of damage from natural background radiation.
See: Self repairing system#In place self-repair
But even there a multi part system is more practical and likely.
So it would not operate alone but rather in conjunction with microcomponent maintenance units.
Microcomponent mainenance units ≠ Molecular assemblers
Main page: Microcomponent maintenance unit
Molecular assemblers are not to confuse with microcomponent maintenance units.
These are also relatively small and compact but they are incapable of mechanosynthesis.
More abstractly they have no manufacturing or demontage capabililities on the lowest assembly level.
Just like a microcomponent recomposer device.
But a microcomponent recomposer device is a macroscopic device whereas a microcomponent maintenance unit is a microscopic one.
Molecular assemblers in the technical book Nanosystems
There aren't any. They are not mentioned.
The closest thing to molecular assemblers in Nanosystems is Section 16.3.6. Minimal diamondoid-material systems and
it starts with: >>Any attempt to describe a "minimal" system must be considered speculative.<<
There is no proposal to build a diamondoid proto-system with a scanning probe tool (of macro- or microscale).
Scanning probe tools are only mentioned (subsection f. Summary.) as tools for analysis:
>>Molecilar probes can be used to determine the outcome of operations within a single device, ...<<
If anything this is rather meant as a later stage in the technical development ladder
that one likely will finds better/other solutions for (e.g. a less compact system).
Also there is no mention about mobility and free-floating non-mountedness.
A trait that is often ascribed to molecular assemblers and
a trait necessary for the grey goo scenario to sound plausible.
Related
- No nanobots
- Mobile robotic device
- Fractal growth speedup limit
- In place assembly and in place mechanosynthesis
- The better alternative that is now instead targeted: Gemstone metamaterial on chip factories
- Still quite compact but less compact self replication by adding one additional assembly level: Second assembly level self replication
This would likely be a system of more or less mobile components on a surface or chip.
(wiki-TODO: add image of dividing cells illustrating the analogy - use it on other related pages too - goo)