Difference between revisions of "Foldamer technology stiffness nesting"
(basic page - likely still many errors) |
(No difference)
|
Revision as of 11:33, 17 June 2024
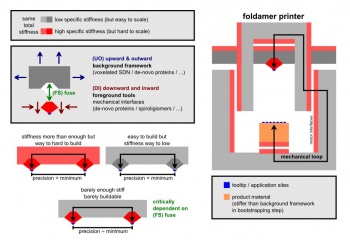
There are various forms of foldamer technology under development.
A pattern that one may spot here is that …
– … the most scalable technologies feature the least stiffness while
– … the least scalable technologies feature the most stiffness
The idea here is to combine the different foldamer technologies to get the best of all the worlds.
More concretely: Positional atomic precision (or near that) is only eventually needed between the workpiece and the tool-tip.
There is no need for positional atomic precision in the much larger "background framework/spacefame".
The background framework merely feature topological atomic precision.
Even with quite a bit of tolerable errors.
The trick here is to look at absolute stiffnesses in N/m
rather than area specific stiffnesses in (N/m)/m² i.e. elastic modulus.
More concretely:
A protein core in a 3D structural DNA background framework matrix features a stuffness not defined solely by the elastic moduli,
but rather by the interface surface area too. This area needs to be factored in to get the total stiffness.
The superatomic wiggles of 3D structral DNA nanotech backround framework that features mere topological atomic precision
maye eventually even statistically cancel out to give positional atomic precision of the nested protein core
and further nested stiff artificial sidechains.
Though achiving all the way to positional atomic precision in one swoop is not strictly necessary.
It's much more likely that things will evolve more gradually.
Maybe this idea is more to treat conceptually.
The incurred challenge
The various different kinds of technologies need to me made compatible in the sence of connectability.
The formable connections need to feature a stiffness that is not notably lower than the stiffness of the
less stiff technology of the pair to connect.
Beside the huge experimental problems with that there's also not much software around for that (as of mid 2024).
The MSEP project has a focus on that cross technology mergability / connectability.
Technologies form least stiff most scalable to most stiff least scalable
- 3D structural DNA nanotechnology - all mere topological atomic precision
- de-novo proteins - positional atomic precision not so floppy side-chains => oringin of fat fingers
- spiroligomers and stiff artificial sidechains - positional atomic precision
- small core crystolecules - if direct path gets so far on it's own - positional atomic precision
Side-note on scalability:
Spiroligomers may be scalable in the quantity of them synthesizable.
Here we look at scalability in therms of selfassembly with termination control though.
Alternate names
- Matryoshka doll stiffness nesting
- Onyon stiffness nesting