Difference between revisions of "Hierarchical selfassembly"
m (→Second assembly level without termination control) |
m (→Related: added * incremental path) |
||
| Line 85: | Line 85: | ||
* [[termination control]] | * [[termination control]] | ||
* ([[topological atomic precision]]) | * ([[topological atomic precision]]) | ||
| + | ---- | ||
| + | * [[incremental path]] | ||
Revision as of 09:22, 5 April 2024
Up: General concept: Convergent assembly
Hierarchical selfassembly (also convergent selfassembly) is the case when
structures get self-assembled from parts that themselves where previously self-assembled.
This implies that hierarchical selfassembly is a subform of iterative self-assembly.
(Iterative self-assembly also covers repeated additions of parts of the same size.)
Note that hierarchical selfassembly is not exclusive to thermally driven self-assembly.
Hierarchical selfassembly is also applicable to:
Contents
Experimental demonstrations
As of time of last review (2024-03) hierarchical selfassembly has been …
- … impressively demonstrated with 3D structural DNA nanotechnology.
- … demonstrated with de-novo proteins
with the large caveat that of missing termination control.
Note that terminating structures like rings or balls is not a form of termination control as defined in this wiki.
Rotation symmetry is an infinite symmetry (just like translation symmetry in a crystal) just that it covers over itself.
Hierarchical selfassembly of 3D structural DNA nanotechnology (3D-SDN)
DNA structures are very soft and mobile thus there is only topological atomic precision present.
But (while merely topological) this already is already real honest to goodness atomically precise LEGO.
At low cryo-temperatures motions may freeze making it more positional atomically precise
Though. 3D-SDN is likely not very compatible with dry conditions in gas or vacuum
likely leading to massive deformations or even fall-apart.
Even individual cryo-frozen water molecules have been imaged by now with stiff cantilever AFM)
3D SDN structures are (just as DNA) not subatomically resolvable with scanning probe microscopy.
Without termination control at the second assembly level
Large (multi micron scale) lattices of 3D-SDN walls have been made by
logical warp-around of encoded positions at the edges surfaces.
This makes for a large super lattice that ate all exact copies of each other and not individually controllable.
(wiki-TODO: Eventually add TEM image from paper showing these walls.)
With termination control at the second assembly level
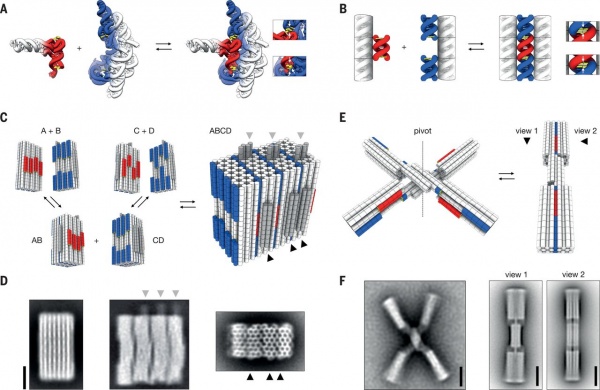
Relevant paper here: (2015) Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components, by Thomas Gerling, Klaus Wagenbauer, Andrea Neuner, Hendrik Dietz
Second self-assembly level for 3D-SDN. Here the assembly and disassembly is (reversibly) controled by salt concentration. The mannequin shape was chosen just to show that shapes can be chosen quite freely. Unlike with cylinders or spherical shell in this case there is some minimal limited amount of termination control present at the second assembly level. One pot reaction. Not sequential.
Voxels at the second self-assembly level
- First level of self-assembly:
Self finding (& folding if long scaffolding strand involved) of the staples strands (short DNA oligomers) to the hexagonal building blocks. Not shown. - Second level of self-assembly:
Self finding of the hexagonal building blocks. Shown.
Here this goes one step further in attempting to make
a standard set of blocks for the second assembly level such
that arbitrary larger structures can be built quickly and easily.
You get individually controllable voxels at the second self-assembly-level too.
Custom blocks may be more versatile, yes, but are one off solutions for specific desired geometries.
(TODO: Check if the following covers termination control. https://link.springer.com/article/10.1557/s43579-022-00248-8)
Hierarchical selfassembly with de-novo proteins
Second assembly level without termination control
First level of self-assembly is self folding of the polypeptide chain.
That might happen right after protein expression.
The big caveat and still present obstacle to overcome here with proteins is absence of termination control.
(wiki-TODO: Add the paper (and TEM image) of non-terminating de-novo protein walls.)
Done't be fooled by the beauty of scale and symmetry of de-novo protein structures that already can't be made.
The difficult part is breaking the symmetry in near arbitrary ways. Like individually controllable voxels.
Related
- Selfassembly level in self assembly
- complementary is: hierarchical positional assembly here
covered by page convergent assembly & assembly level in positional assembly